Gene flow estimation and mismatch distribution indices of Donkeys based on MtDNA
Abstract
A total of 20 donkeys were used for the study. The animals were selected using the criteria of unrelated individuals, samples from different genetic groups and household in order to reduce the genetic relationship among animals and to increase the breed representativeness. Blood samples was collected through the jugular vein and immediately transferred into EDTA bottles. All samples were kept at 4 °C until the further laboratory process. The DNA extraction, PCR and gel electrophoresis were done in DNA laboratory in Kaduna (DNA lab off kinkino road unguwanRimi) Kaduna State. A total of 16 sequences from the present study and together with 66 sequences from the gene bank were used for the above analysis. The results received were blast in national center for biotechnology information (NCBI) in other to ascertain their purity. Bio edit program was used to check and edit ambiguous bases. The program ClustalW in MEGAX was used for multiple sequence alignments. Numbers of haplotypes, numbers of nucleotide polymorphic sites, haplotype diversity (h), and nucleotide diversity (π) were analysed with DnaSPver 6.0, haplotype joining network was constructed using Median Joining Network in NETWORK v 10.1 and the AMOVA for nucleotide variance using Arlequin software version 3.5. The gene flow indicates migration among donkey population in the study area. The non - significant Fst value obtained in the present study indicates absence of phylogeographic sub- structure in the study area. The negative Tajima D may be suggestive of population expansion or purifying selection which implies that such populations are not at equilibrium. The phylogenetic relationship indicates that the studied donkeys were closely related to Equus zebra (zebra), Equus ferus (horse), Equus quagga burchellii (British zebra), Equus asinus africanus (Wild ass) and Equus kiang (Asinus of Tibetan plateau) respectively.
Introduction
Donkey (Equus asinus), occurs throughout most of semi-arid Africa today, much of its distribution is still spreading in eastern and southern Africa. The ass was probably domesticated in northeast Africa. It seems to have spread to Sub-Saharan West Africa relatively late (Epstein, 1984; Groves, 1986: Eisenmann, 1999; Blench, 1995). Although there are donkeys in villages throughout the semi-arid north, their susceptibility to internal parasites and trypanosomiasis restrict their all year-round use in the more humid southern states. According to Fielding and Krause (1998), donkeys adapt well to hot-dry desert environment through body temperature control, water metabolism, and special nutritional and anatomic features. Donkeys in the tropics are able to maintain homoeothermic by compensatory mechanisms in order to keep their physiological values within the established normal range (Minka and Ayo, 2007). The most limiting factor for survival in semi-arid and arid areas is during drought, that is, when water is not available. Donkeys survive better than cattle in these areas and during drought (Smith and Pearson, 2005). They have the ability to tolerate thirst and this allows them to have access to more remote source of forage, inaccessible to cattle in rangeland (Smith and Pearson, 2005). Nengomasha et al. (1999) reported that donkeys with limited access to water (2 to 3 days) loose less water through faeces than their counterparts with ad libitum access to water. This is because faecal water loss can account for 50% of all water lost from the body. Donkeys are also adapted to low-quality, high-fibre feed, which contributes to their ability to eat and survive on very little quantity of feed (Nengomasha et al., 2000). The gene flow estimation recorded the haplotype data information and the sequence data information. The genetic coefficient of differentiation (Gst) which is the estimate that measure the genetic differentiation is inversely proportional to the gene flow.
Material and Methods
Location of the study
This study was conducted in the Sahel agro-ecological zone of Nigeria. The Sahel agro-ecological zone comprises the following states in North East and some state in the North West of Nigeria: Borno, Yobe, Kano, Katsina and Sokoto. The Sahel ecological zone is characterized by vast grassland and few trees. The temperature ranges from 33 oC to 40 oC and humidity percent ranging from 4 - 12% with annual average rainfall of 400 – 600mm. The agricultural activities in the area include arable crop farming, livestock rearing, fishing and hunting. This study was carried out specifically in Borno and Yobe State north eastern part of Nigeria. Borno State is located between latitude 10 and 14 oE and Longitude 11 and 14o N and an altitude of 354 m above sea level. It covers an area of 61,435 km2 which is about 12% of the total area of the country. It occupies a greater part of the Chad Basin and shares border with Adamawa State to the south-east, Gombe State to the south west and Yobe State to the north-west (BOSHIC, 2007).
Animal and DNA isolation
A total of 20 donkeys were used for the study of molecular characterization. The animals were selected using the criteria of unrelated individuals, samples from different genetic groups and household in order to reduce the genetic relationship among animals and to increase the breed representativeness. Blood samples was collected through the jugular vein and immediately transferred into EDTA bottles. All samples were kept at 4 °C until the further laboratory process. The DNA extraction, PCR and gel electrophoresis were done in DNA laboratory in Kaduna (DNA lab off kinkino road unguwanRimi) Kaduna state.
Data analysis
The results received were blast in national center for biotechnology information (NCBI) in other to ascertain their purity. Bio edit program was used to check and edit ambiguous bases. The program ClustalW in MEGAX was used for multiple sequence alignments. Numbers of haplotypes, numbers of nucleotide polymorphic sites, haplotype diversity (h), and nucleotide diversity (π) were analysed with DnaSPver 6.0, haplotype joining network was constructed using Median Joining Network in NETWORK v 10.1 and the AMOVA for nucleotide variance using Arlequin software version 3.5. A total of 16 sequences from the present study and together with 66 sequences from the gene bank were used for the above analysis.
Results
Gene flow estimation
Table 1 presents the gene flow from estimation using the haplotype data information. The genetic coefficient of difference gene obtained in the study Gst was 0.12221 while the effective population number Nm (number of migrants) was 1.80. The estimation of gene flow using sequenced data recorded the Delta St: 0.02911 Gamma St: 0.58661 and the Nm: 0.18. The measures of nucleotide sub-division Nst: 0.58010 and Nm: 0.18 and the population sub- division Fst: 0.58010.
Table 1: Gene Flow Estimation
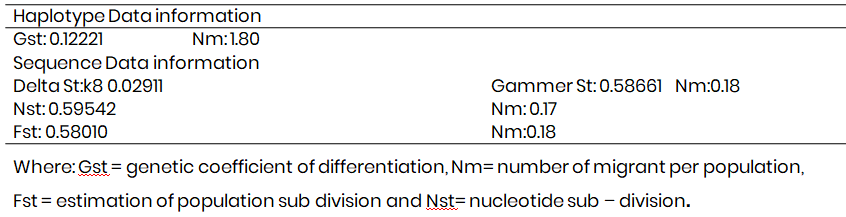
Mismatch distribution indices of the mtDNA of donkeys in Sahel agro ecological zone of Nigeria
Table 2 presents the mismatch distribution indices of the mtDNA of donkeys. The results recorded negative Tajima D (-0.1114) and Harpendings Raggedness index value of (1.111) while the mutation parameters were highest ϴ0 and ϴ1 with the values of 70.398 and 7366.36 respectively. Fig. 1 presents the population structures of the donkey.
Table 2: Mismatch distribution indices of the mtDNA of Donkeys in Sahel agro - ecological zone of Nigeria
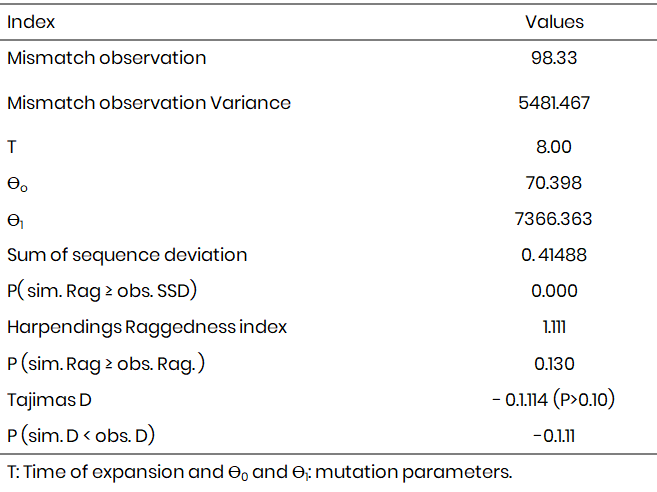
Analysis of molecular variance (AMOVA)
Table 3 presents the analysis of Molecular variance. The results indicated that 63.48 % of the variation was among the maternal genetic difference in the study area. Only 27.79 % variance was within the sample country population while 8.73 % was obtained as the among group variation in the study area. The Fst obtained was not significant in the study Fst = 0.132. Fig. 3 present the population structures of donkey in the study area.
The phylogenetic tree showing the relationships between Borno/Yobe donkeys is shown in Fig. 4. The results grouped donkeys into three clades, clade A consists of donkeys from other regions (Indian, Cameroon, China, Pakistan, Turkey, Korea etc) with White Borno donkeys. Clade B grouped donkeys into (Black Borno. Black Yobe and Red/rust donkeys) while clade C grouped donkeys of (Red/rust Borno, White Yobe, Grey Yobe, Black Borno and White Yobe donkeys) in its clustered. The out grouped was Gallus- gallus.
Different populations were distinguished by use of colour codes (Red = Borno, Yellow =Yobe, purple = India, Light Blue = Cameroon, Pusher pink = India, Light Green = India, Light Purple = Turkey, Red = Pakistan donkey and Green = India). Area of each circle is proportional to the frequency of the corresponding haplotype(s) Fig. 2.
Neighbour - joining tree constructed with the sum of branches length of 1.47044117 between the studied donkey and other equine species. The number at the node represents the percentage bootstraps values for interior branches after 1000 replication.
Fig. 3 Presents the phylogenetic tree of donkeys and other Equine species. The results reveals that the red/rust donkeys in Borno were the most ancient breeds among the studied donkeys followed by the (White and Black Borno donkeys and White and Grey Yobe donkeys) and lastly followed by the (Black and grey donkeys Borno and red/rust donkeys in Yobe). The evolutionary tree discovered that donkeys were closely related with Zebra (Equine zebra) followed by horses (Equine ferus), followed by British zebra (Equine quagga burchellii), Wild ass (Equine asinus africanus), largest Asinus Tibetan plateau (Equine kiang), zebra in Kenya and Ethiopia and lastly the Imperial zebra (Equine grevyi).
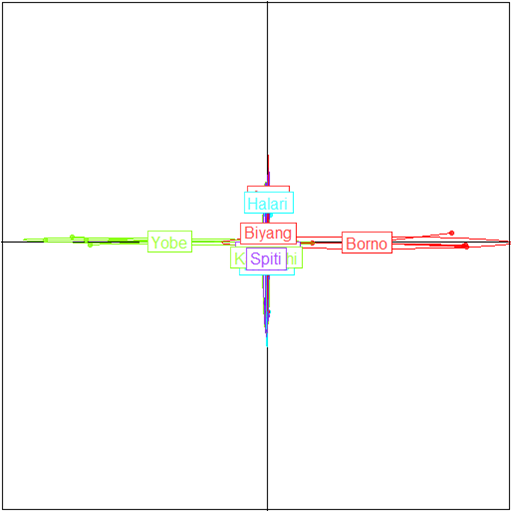
Fig 1: Population structures of the donkeys in Sahel agro ecological zone of Nigeria. The population structures figure 1 was analy
Table 3: Analysis of molecular variance within and among Donkey populations using mtDNA sequences in the study area
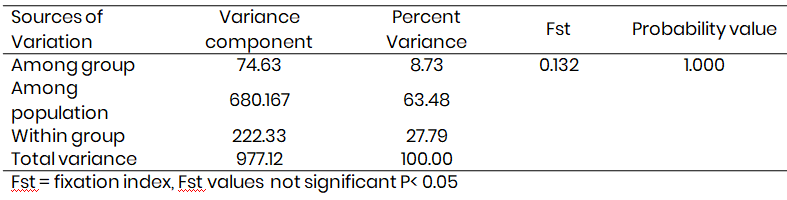
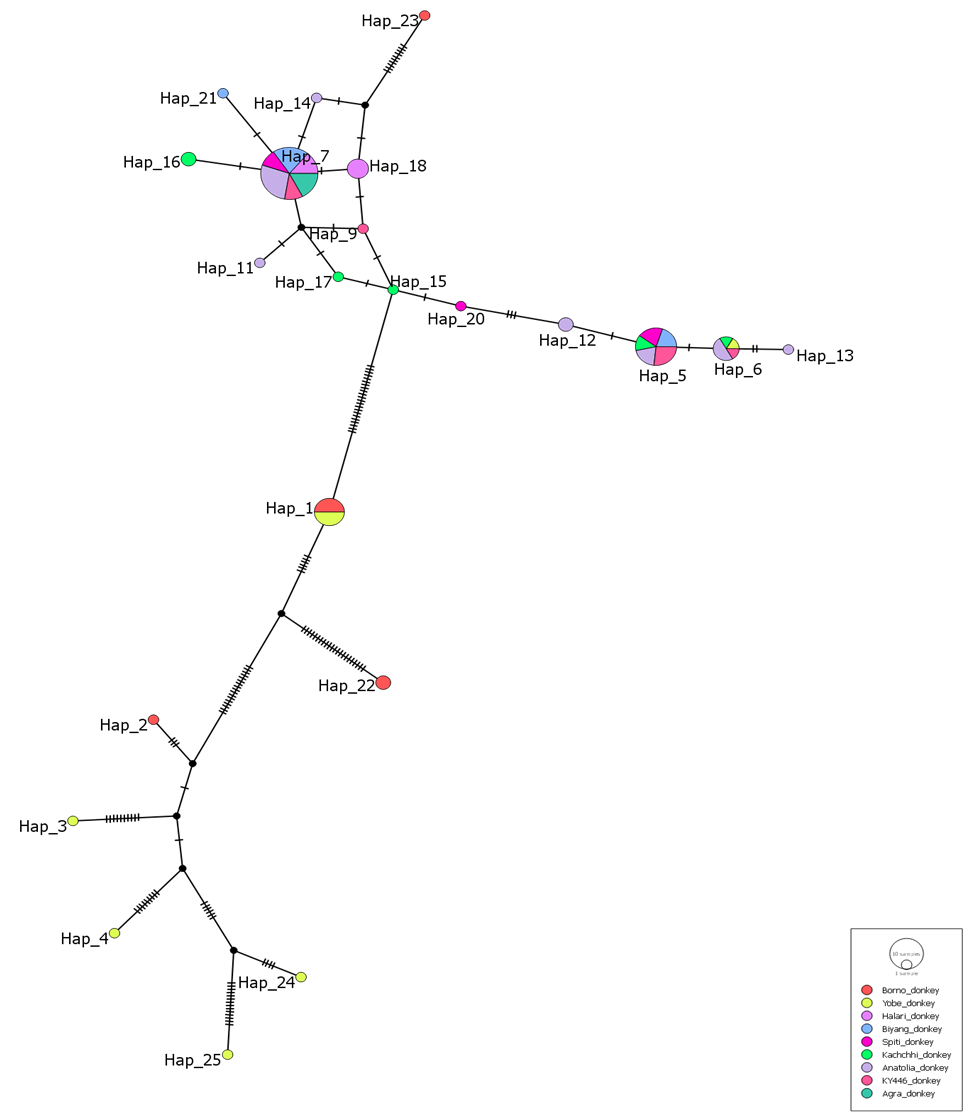
Fig. 2: Median – joining network result for the relationship between the Borno and Yobe donkeys and other population in the
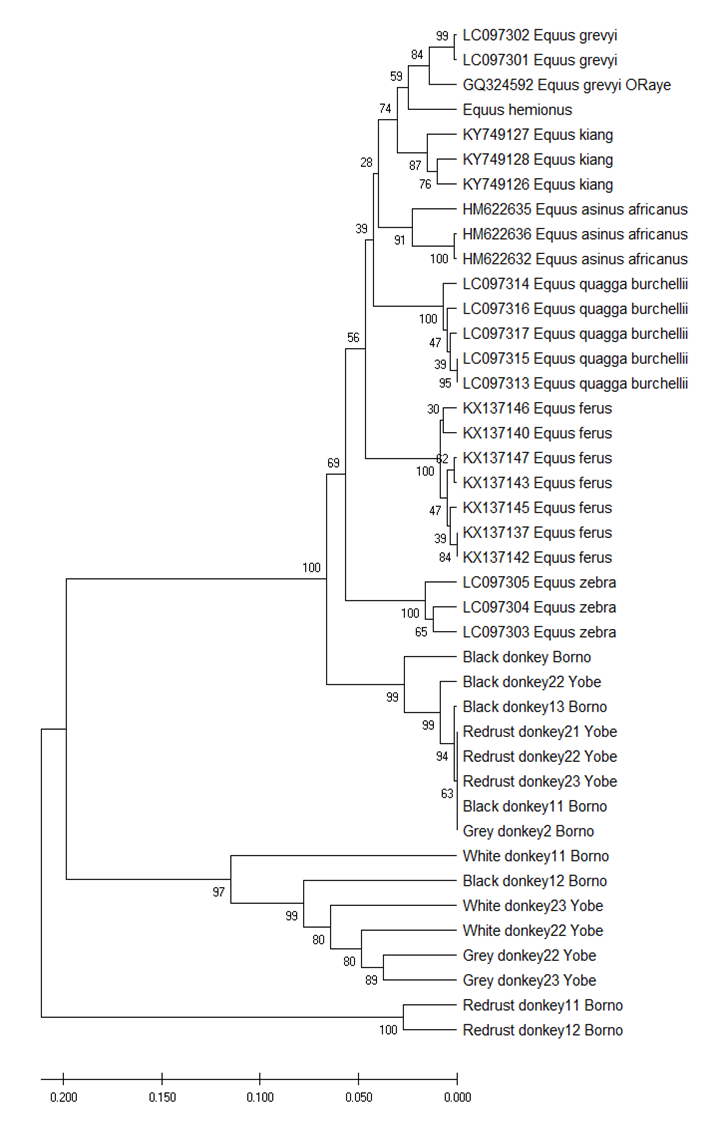
Fig. 3: Neighbour - joining phylogenetic tree reconstructed for donkeys in Sahel agro- ecological zone of Nigeria with other Equin
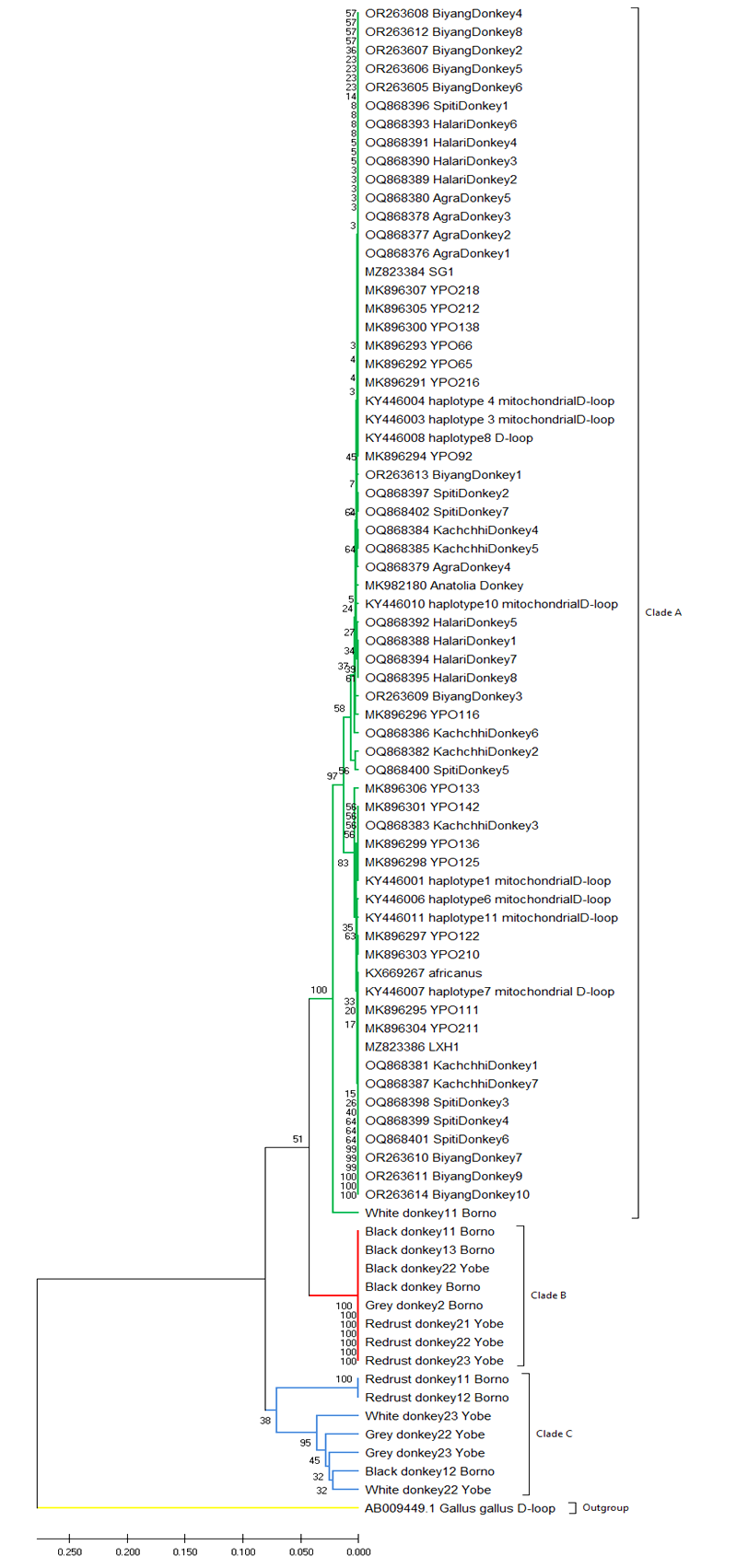
Fig. 4: Neighbour - joining phylogenetic tree reconstructed for donkeys in Sahel agro- ecological zone of Nigeria with other popul
Discussion
Gene flow estimation
The gene flow estimation recorded the haplotype data information and the sequence data information. The genetic coefficient of differentiation (Gst) which is the estimate that measure the genetic differentiation is inversely proportional to the gene flow. The Gst ranges from 0.0 - 1.0 as expected, with 0.0 represents no difference in allele frequency between two populations and 1.0 indicating the two population are fixed for alternate allele. The gene flow recorded migrates among donkey population in the study area which is in line with some authors (Hedrick, 2005; Meirmans and Hedrick, 2011 and Gupta et al., 2018). The estimation of population sub - division (Fst) and the nucleotide sub – division (Nst) were used to measure genetic distance and were generalized for multiple alleles. Both values obtained were low. This means that the measure of the difference in the allele frequency of these gene between the populations were low. The presents study agreed with the findings of Anila et al. (2014), Agaviezor et al. (2017) stated that if population are similar in size then Nm (number of migration per population) describes the average number of individual per generation migrating between population. The Island model by Wright (1969) determined Nm theoretically that if more than one individual migrates between populations every other generation (Nm> 0.5), different alleles at a locus in the two populations would not become fixed due to genetic drift. The major determination of population structures when Nm > 0.05 and significant genetic differentiation can result from gene drift when Nm < 0.5 (Wolf and Soltis, 1992).
Mismatch distribution and Test of Neutrality
The mismatch distribution indicates the genetic difference between sample and the population growth. Although it indicates population expansion it does not affect the population structures among the samples (Harpending et al., 1994). The negative Tajima D results suggested population expansion or purifying selection (Tajima, 1996), indicating that such population are not at equilibrium nor experiencing random (neutral) selection. Negative Tajima D was also observed among Halari donkeys (Bhardwaj et al., 2012). Hahn et al. (2002) reported that such population tended to have excess of rare alleles. The present findings of Borno and Yobe donkeys investigated that they were not at neutral equilibrium. This is in line with (Harpending, 1994; Rogers, 1995; Schneider and Excoffier 1999; Jobling et al., 2004). These types of populations conformed to an estimated sudden demographic model given the mutation parameters (since t >0 and ϴ1> ϴ0). The non - significant Fst value obtained in the present study indicate there was no population structure in the donkey population.
Phylogenetic relationships between Donkeys and other equine species
The present results indicate that the studied donkeys were closely related to zebra, horse, wild ass and the imperial zebra, respectively. This agrees with some other researches who revealed that the donkey and the horse share common ancestors approximately 6.4–12.7 million years ago (Waddell et al., 1999; Nikido et al., 2001 and Huang et al., 2015). The phylogenetic relationship showed that Nubian lineages had made a genetic contribution to the evolution of the donkey of Sahel agro-ecological zone of Nigeria indigenous donkey evolution as investigated by Earnist et al. (2021). The Equus asinus africanus (wild ass) is most closely related to the donkey, and together they form a sister group with the horse. This finding did not agree with the present results which discovered that the Nigerian donkey is closely related with zebra (Waddell et al., 1999 and Nikido et al., 2001). Horses, donkeys and zebras belong to the genus Equus, which diverged approximately 4 – 4.5 million years ago (Orlando et al., 2013). The ancestors of today donkeys and zebras dispersed between 2.1 and 3.4 million years into an American continent, eventually experiencing major population expansion and collapse that coincide with past climate change event and this agrees with the present results. These promoted equids as a fundamental model for understanding the inter play between chromosomal structure, gene flow and eventually species formation (Orlando et al., 2013).
References
Agaviezor, B. O., Ajayi, F. O., & Omayiho, T. W. (2017). Bone morpho-genetic protein 3 (BMP3) gene variation in some livestock animals. Nigerian Journal of Biotechnology, 32, 28-32.
Anila, H., Yili, B., & Petrit, D. (2014). Genetic diversity of Albanian goat by mtDNA sequence variation. Nigerian Journal of Biotechnology, 14, ISSN: 1310-2818 (print), 1314-3530.
Bhardwaj, A., Pal, Y., Legha, R. A., Sharma, P., Nayan, V., Kumar, S., Tripathi, H., & Tripathi, B. N. (2020). Donkey milk composition and its therapeutic application. Indian Journal of Animal Science, 90(6), 837-841.
Blench, R. M. (1995). A history of domestic animals in Northeastern Nigeria. Cahiers de Science Humaine, 31(1), 181-238. ORSTOM.
Borno State Ministry of Home Affairs Information and Culture (BOSHIC). (2007). Borno state ministry of home affairs information and culture. Retrieved from http://www.bornonigeria.com /index.phoption.com(Accessed 20 March 2017)
Earnist, S., Nawaz, S., Ullah, I., Bhinder, M. A., Imran, M., Rasheed, M. A., Shehzad, W., & Zahoor, M. Y. (2021). Mitochondrial DNA diversity and maternal origins of Pakistani donkeys. Journal of Brazilian Biology, 1-6. ISSN: 1519-6984.
Eisenmann, V. (1999). L’Origine des ânes : questions et réponses paléontologiques. Ethnozootechnie, 56, 5-26.
Epstein, H. (1984). Ass, mule and onager. In I. L. Mason (Ed.), Evolution of Domesticated Animals (pp. 174-184). London: Longman.
Fielding, D., & Krause, P. (1998). Physiology, nutrition and feeding. In D. Fielding & P. Krause (Eds.), Donkeys (pp. 14-31). London: MacMillan Education Ltd.
Groves, C. P. (1986). African wild ass (Equus africanus) and domesticated donkey (Equus asinus): Fact sheet: Taxonomy and history.
Copyright
Open Access: This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.

