CRISPR-Cas revolution in Agriculture: From precision genome editing to sustainable crop improvement
Abstract
The CRISPR-Cas system has emerged as a transformative tool in agricultural biotechnology, revolutionizing the landscape of crop improvement. This review paper explores the multifaceted applications of CRISPR technology in agriculture, from its fundamental principles to its practical implementations for precision genome editing. We delve into the various strategies employed to enhance crop growth and yield traits, including disease resistance, abiotic stress tolerance, and nutritional content, thereby contributing to the development of sustainable agriculture practices. Furthermore, we discuss the regulatory frameworks and ethical considerations surrounding the deployment of CRISPR-edited crops, highlighting the challenges and opportunities for its widespread adoption. Through a comprehensive analysis of recent advancements and future prospects, this review aims to provide insights into the role of CRISPR-Cas in shaping the future of agriculture and global food security.
Introduction
The application of CRISPR-Cas9 technology in agriculture has catalyzed a paradigm shift in crop improvement strategies, offering unparalleled precision and efficiency in genome editing. CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and its associated protein Cas9 constitute a groundbreaking molecular toolkit that enables targeted modifications of DNA sequences within plant genomes (Jinek et al., 2012). This revolutionary genome editing platform has not only expedited the pace of genetic manipulation in crops but also unlocked novel avenues for sustainable agriculture and global food security (Hsu et al., 2014).
The versatility and simplicity of the CRISPR-Cas9 system have propelled research efforts worldwide, driving significant advancements in crop biotechnology. By precisely targeting genes linked to agriculturally significant traits, such as yield, stress tolerance, and nutritional composition, researchers have achieved remarkable success in developing crops with enhanced productivity and resilience (Puchta and Fauser, 2014). For instance, CRISPR-mediated editing has been instrumental in conferring resistance to devastating pathogens like powdery mildew in wheat (Wang et al., 2014), bacterial blight in rice (Li et al., 2012), and citrus canker in citrus plants (Jia and Wang, 2014).
Moreover, CRISPR technology holds promise for promoting sustainable agricultural practices by mitigating the environmental impacts associated with conventional farming methods. Through targeted genome editing, researchers can engineer crops that require fewer chemical inputs, such as pesticides and fertilizers, thereby reducing environmental pollution and preserving soil health (Schindele et al., 2018). Additionally, CRISPR-edited crops with improved nutritional profiles offer a viable solution to malnutrition and food insecurity, particularly in regions where staple crops lack essential vitamins and minerals (Wurtzel et al., 2019).
Despite the transformative potential of CRISPR-Cas9 in agriculture, several challenges persist, ranging from regulatory complexities to ethical considerations and public acceptance (Waltz, 2016). The regulatory landscape governing the cultivation and commercialization of genetically modified organisms (GMOs) varies across jurisdictions, presenting obstacles to the global adoption of CRISPR-edited crops (Chawla et al., 2017). Moreover, concerns regarding unintended off-target effects and the potential for gene flow to wild relatives underscore the need for rigorous risk assessment and environmental monitoring (Bortesi and Fischer, 2015).
In this comprehensive review, we aim to provide an extensive exploration of the CRISPR-Cas revolution in agriculture, spanning its diverse applications, underlying mechanisms, regulatory frameworks, and ethical implications. By synthesizing recent research findings and emerging trends, we seek to elucidate the transformative potential of CRISPR technology in driving sustainable crop improvement and addressing the multifaceted challenges confronting modern agriculture.
CRISPR-Cas: components and mechanism
Components of CRISPR-Cas system
At its core, the CRISPR-Cas system consists of two primary components: the Cas protein and the guide RNA (gRNA). The Cas protein, typically Cas9, serves as the molecular scissors responsible for cleaving DNA at specific target sequences. The gRNA, composed of a CRISPR RNA (crRNA) and a trans-activating CRISPR RNA (tracrRNA) fused together, guides the Cas protein to the target DNA sequence through complementary base pairing (Jinek et al., 2012).
Mechanism of CRISPR-Cas action
The mechanism of CRISPR-Cas-mediated genome editing involves several key steps. Initially, the gRNA forms a complex with the Cas protein, leading to the formation of the Cas-gRNA ribonucleoprotein (RNP) complex. This complex scans the genomic DNA for sequences complementary to the gRNA, facilitating target recognition (Doudna and Charpentier, 2014).
Upon binding to the target DNA sequence, the Cas protein undergoes a conformational change, resulting in the activation of its endonuclease activity. The endonuclease domains of the Cas protein then catalyze the cleavage of the DNA, generating double-strand breaks (DSBs) at the target site (Gasiunas et al., 2012).
Following DNA cleavage, the cell's DNA repair machinery comes into play to resolve the DSBs. Two primary pathways involved in DNA repair are non-homologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ often leads to small insertions or deletions (indels) at the site of the DSB, resulting in gene knockout or disruption. In contrast, HDR utilizes a template DNA molecule to precisely repair the DSB, enabling gene editing and insertion of desired sequences (Doudna and Charpentier, 2014).
CRISPR-Cas9: evolution to precision genome editing
The journey of the CRISPR-Cas system began with the elucidation of clustered regularly interspaced short palindromic repeats (CRISPR) in the genomes of bacteria and archaea. Initial studies in the late 1980s and 1990s identified these repetitive DNA sequences, which sparked curiosity about their function. However, it was not until the early 2000s that researchers began to unravel the significance of CRISPR in bacterial immunity against viral infections.
The breakthrough came in 2012 when Doudna and Charpentier demonstrated the programmable nature of CRISPR-Cas9 for targeted genome editing in bacteria. Their landmark paper published in Science described how the Cas9 protein, guided by a short RNA molecule, could precisely cleave specific DNA sequences. This pivotal discovery laid the foundation for a myriad of applications in genetic engineering, ranging from gene knockout and knock-in to gene regulation and functional genomics.
The versatility of CRISPR-Cas9 lies in its ability to target virtually any genomic locus by simply modifying the sequence of the gRNA. This programmable nature, coupled with its high efficiency and specificity, has revolutionized genome editing and facilitated the study of gene function and regulation in various organisms.
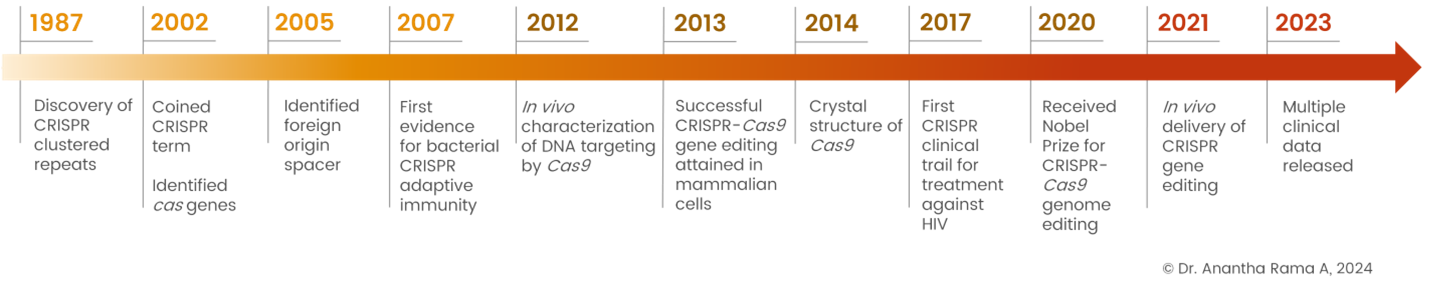
Fig. 1: Evolution of CRISPR-Cas system: Key Milestones
Importance of CRISPR-Cas in Agriculture
The escalating impacts of climate change have intensified the specter of food scarcity, necessitating innovative approaches to bolster agricultural productivity. Climatic shifts, characterized by erratic weather patterns, extreme temperatures, and unpredictable precipitation, have destabilized traditional farming practices, leading to reduced crop yields and compromised food security. In response to these challenges, targeted genome editing, facilitated by advanced molecular tools such as CRISPR-Cas systems, has emerged as a promising approach to boost agricultural productivity. In rice and wheat, targeted genome editing has been instrumental in increasing grain size, weight, and number, as well as enhancing protein content, tiller spread, and tiller number. These improvements have been reported in various studies (Wang et al., 2020; Oliva et al., 2019; Zhang et al., 2019).
Moreover, targeted genome editing has led to significant enhancements in the quality of crops such as rice and corn. Modified crops utilizing the CRISPR–Cas system have been tailored to reduce the levels of toxic steroidal glycoalkaloids, thereby enhancing the color and extending the shelf-life of fruits and vegetables, rendering them more commercially appealing. Additionally, these modifications have resulted in an increase in desirable traits such as amylose and starch content, as well as good fats like oleic acid levels. Furthermore, improvements in fragrance, gluten protein reduction, and decreased unsaturated fatty acids content have been achieved (Li et al., 2012; Cermak et al., 2015; Clasen et al., 2016; Jia et al., 2017). These advancements highlight the transformative potential of CRISPR-based targeted genome editing in agriculture, offering precise and tailored solutions to address the complex challenges posed by climate change and food insecurity.
Enhancing crop yield and quality
CRISPR-Cas9 genome editing, targeting the OsNAS2 promoter, specifically deleting the cis-regulatory element ARR1AT at position -933, significantly increased Zn concentration per plant in rice and also led to an augmented spikelet number per main panicle, resulting in increased grain yield per plant (Ludwig et al., 2024). In an another study conducted by Usman et al. (2020) reported that precise editing of the OsPYL9 gene by RNA-guided Cas9 nuclease increased the grain yield in rice by regulating circadian rhythm. CRISPR/Cas9-mediated multiplex genome editing targeted three key genes - GW2, GW5, and TGW6, known as negative regulators of grain weight and the outcomes demonstrated a notable increase in grain size and thousand grain weight (Xu et al., 2016).
Bioactive compounds, characterized as additional nutritional constituents found in small quantities in foods, often contribute to the prevention of cardiovascular disease and cancer. Anthocyanin, malate, γ-aminobutyric acid (GABA), and lycopene are among these bioactive compounds. Utilizing CRISPR-Cas9 technology, researchers have enhanced the levels of anthocyanin, GABA, and lycopene in tomato fruits by modulating the expression of key genes in their metabolic pathways (Cermak et al., 2015; Nonaka et al., 2017).
The function of TM6 in strawberry was elucidated using the CRISPR-Cas9 system applied to an octoploid species. Phenotypic analysis of tm6 mutants unveiled pronounced defects in anthers, underscoring TM6's crucial role in flower development (Martín-Pizarro et al., 2019). Furthermore, CRISPR-Cas9 was employed to explore the biological role of YUCCA 10 (YUC10) in auxin synthesis during strawberry fruit development. Knocking out YUC10 resulted in a significant reduction in free auxin in yuc10 mutants (Feng et al., 2019).
Disease resistance
CRISPR–Cas13a presents an efficient tool for targeting RNA viruses, predominantly plant viruses. Aman et al. 2018 utilized LshCas13a to target Turnip mosaic virus (TuMV) which cause Turnip mosaic disease in Nicotiana benthamiana, achieving significant reductions in viral gene expression. The predominant approach for pathogen control via the CRISPR/Cas9 system involves disrupting the host's susceptibility gene (S gene), thus impeding plant-pathogen interactions and preventing pathogen establishment (Zaidi et al., 2018). This disruption can be achieved by targeting either the promoter sequence of the S gene or interrupting the effector-binding site. Ali and his coworkers effectively demonstrated virus targeting by inducing indels in the genome of tomato yellow leaf curl virus, thus imparting viral resistance. This resistance was achieved through CRISPR/Cas9 binding to the viral genome, subsequently obstructing the viral genome’s access to replication units, or by generating blunt-end cuts or indel mutations on the viral genome. Thomazella et al. (2016) utilized the CRISPR-Cas9 system to deactivate the DMR6 ortholog in tomatoes. The resulting dmr6 mutants exhibited disease resistance against a range of pathogens, such as Pseudomonas syringae, Phytophthora capsica, and Xanthomonas spp., with minimal adverse effects. Pseudomonas syringae induces bacterial speck disease in tomato plants, which significantly impacts their productivity and market value. Given the role of Jasmonatezim domain protein 2 (JAZ2) in defense against P. syringae in A. thaliana, scientists employed CRISPR-Cas9 to produce dominant JAZ2 repressors in tomatoes with the C-terminal jasmonate associated (Jas) domain removed (JAZ2Δjas). These JAZ2Δjas repressors confer resistance to P. syringae. Nekrasov et al. (2017) employed CRISPR-Cas9 technology to create a tomato loss-of-function mlo1 mutant. This mutant exhibited complete resistance to the powdery mildew fungus Oidium neolycopersici.
Herbicide resistance
The application of the CRISPR-based gene editing technique has led to the successful development of crop varieties resistant to herbicides that target the ALS enzyme. This technique has been implemented across various crops, such as rice (Zhang et al., 2021), maize (Li et al., 2020), wheat (Zhang et al., 2019), watermelon (Tian et al., 2018), oilseed rape (Wu et al., 2020), tobacco (Kang et al., 2019), tomato and potato (Veillet et al., 2019). Additionally, wheat has shown tolerance to herbicides inhibiting ACCase through cytidine-deaminase-mediated base editor (CBE). To enhance the efficiency of CRISPR/Cas technology, the target-activation induced cytidine deaminase (Target-AID) system has been introduced, facilitating the simultaneous improvement of multiple traits in crops.
In the development of herbicide-resistant crop varieties, only resistance to ALS-inhibiting herbicides, ACCase-inhibiting herbicides, and glyphosate has seen significant success. However, research on the widespread adoption and effective management of weeds with herbicides that target 4-hydroxyphenyl pyruvate dioxygenase and protoporhyrinogen oxidase is lacking.
Plant stress resistance
Stress poses a formidable challenge to agricultural productivity, with abiotic and biotic stressors exerting detrimental effects on crop yield. Abiotic stressors, encompassing factors such as drought, floods, temperature extremes, salinity, heavy metals, and radiation, disrupt plant growth and development. Conversely, biotic stress arises from attacks by various pathogens including viruses, bacteria, fungi, and herbivores, further compromising crop health and productivity. To mitigate these challenges, crops such as rice, tomato, cucumber, and grapefruits have been genetically modified through induced mutations to enhance resistance to both abiotic (Klap et al., 2017) and biotic stresses (Lu et al., 2018). While earlier attempts at site-specific genomic mutation relied on DNA-binding endonucleases such as zinc finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN), these approaches have inherent limitations (Christian et al., 2010). The advent of the CRISPR–Cas system marked a significant breakthrough, enabling precise genome editing in a wide range of crops including rice, wheat, Nicotiana benthamiana, and Arabidopsis (Chen et al., 2019). In their study, Li et al. (2018) discovered that C-repeat binding factor 1 (CBF1) plays a crucial role in safeguarding plants against cold injury. The cbf1 mutant, created using CRISPR-Cas9, displayed exacerbated chilling-injury symptoms with increased electrolyte leakage compared to wild-type (WT) plants. Additionally, MAPK3, known for its involvement in resisting gray mold disease (Zhang et al., 2018), also contributes to tomato drought response by shielding cell membranes from oxidative damage.
Customized sgRNA-Cas9 systems have emerged as a widely employed tool for genome modification in crops like rice and wheat, showcasing the ease and efficiency of genome editing (Shan et al., 2013). Notably, Cas12a, formerly known as Cpf1, presents advantages over Cas9 in plant genome editing due to its requirement of shorter guiding nucleotides, ability to create larger deletions at target sites, and facilitation of NHEJ-mediated donor DNA insertion (Kim et al., 2017).
In Arabidopsis, Feng and coworkers successfully demonstrated the mutation and heritability of five endogenous target genes – brassinosteroid insensitive 1 (bri1), jasmonate-zim-domain protein 1 (jaz1), gibberellic acid insensitive (gai), magnesium chelatase subunit i (chli), and transparent testa 4 (tt4), in addition to the apetala1 (ap 1) gene, using CRISPR–Cas tools (Feng et al., 2014). Furthermore, CRISPR–Cas technology can be harnessed for the regulation of genes responsible for epigenetic modification, methylation, and/or demethylation, enabling simultaneous induction and repression of gene expression (Puchta, 2016).
Hybrid breeding, alongside precision plant breeding facilitated by CRISPR–Cas, holds promise for increasing crop productivity (Chen et al., 2019). CRISPR–Cas has been instrumental in producing thermosensitive male-sterile lines in rice (Zhou et al., 2016) and maize (Svitashev et al., 2016), facilitating the production of high-quality hybrid varieties. Additionally, knockout mediated by CRISPR–Cas has enabled the development of herbicide-resistant crops in rice (Shimatani et al., 2017), Arabidopsis (Chen et al., 2017), and watermelon (Tian et al., 2018), further expanding the scope of genome editing applications in plants.
|
Crops |
Targeted gene |
Result |
References |
|
|
Agricultural crops |
||||
|
Rice |
OsSEC3A, OsSWEET13, OsERF922 |
Resistant to blast and bacterial blight |
Ma et al., 2018 |
|
|
Rice |
ALS |
Herbicide resistance |
Chen et al., 2019 |
|
|
Rice |
UVb1-1 |
Resistant to false smut |
Mishra et al., 2018 |
|
|
Rice |
OsGS3 |
Increase in grain size |
Miao et al., 2013 |
|
|
Wheat |
EDR1 |
Resistant to powdery mildew |
Zhang et al., 2017 |
|
|
Barley |
ENGase, HvPM19, BolC.GA4.a |
Increase in number of grains |
Kapusi et al., 2017 |
|
|
Maize |
ARGOS8 |
Drought resistance |
Svitashev et al., 2016 |
|
|
Horticultural crops |
||||
|
Tomato |
SlMLO1 |
Resistant to powdery mildew |
Nekrasov et al., 2017 |
|
|
Potato |
ALS |
Herbicide resistance |
Choudhury et al., 2016 |
|
|
Cucumber |
eIF4E |
Broad virus resistant |
Sauer et al., 2016 |
|
|
Apple |
DIPM1, DIPM2, DIPM4 |
Resistant to fire blight disease |
Malnoy et al., 2016 |
|
|
Kiwifruit |
CEN4, CEN |
Rapid flower and fruit development |
Varkonyi et al., 2018 |
|
|
Grape |
VvMLO3 |
Resistant to powdery mildew |
Wan et al., 2020 |
|
|
Citrus |
CsLOB1 |
Resistant to citrus canker |
Jia et al., 2017 |
|
|
Cocoa |
TcNPR3 |
Resistant to Phytophthora tropicalis |
Fister et al., 2018 |
|
|
Watermelon |
ALS |
Herbicide resistance |
Tian et al., 2018 |
|
|
Papaya |
alEPIC8 |
Resistance to Phytophthora palmivora |
Gumtow et al., 2018 |
|
|
Cassava |
EPSPS |
Herbicide resistance |
Hummel et al., 2018 |
|
|
Soybean |
GmSPL9a, b, c |
Increase in yield |
Bao et al., 2019 |
|
|
Mushroom |
PPO |
Browning resistant |
Waltz, 2016 |
|
CRISPR crop regulations and ethics
The regulatory framework surrounding CRISPR-modified crops varies significantly among countries and regions. In some jurisdictions, CRISPR-edited crops that do not involve the insertion of foreign DNA are subject to less stringent regulations compared to traditional genetically modified organisms (GMOs). For example, the European Union (EU) has classified some CRISPR-edited crops as non-GMOs, thereby exempting them from rigorous regulatory requirements (Eckerstorfer et al., 2019).
In contrast, other countries, such as the United States, have adopted a case-by-case approach to regulate CRISPR-modified crops, evaluating them based on their characteristics and potential risks to human health and the environment. The U.S. Department of Agriculture (USDA), the Environmental Protection Agency (EPA), and the Food and Drug Administration (FDA) play key roles in assessing the safety and environmental impact of CRISPR-modified crops (Waltz, 2018).
Ethical Implications
CRISPR-mediated genome editing in agriculture raises various ethical considerations that must be carefully addressed. One of the primary concerns revolves around unintended consequences and potential ecological impacts of genetically modified crops. Altering genes in crops could inadvertently affect ecosystems, biodiversity, and non-target organisms, leading to unforeseen environmental consequences (Lassoued et al., 2019).
Additionally, ethical considerations extend to issues of social justice and equity in access to CRISPR technology and its benefits. There is a risk that CRISPR-based agricultural innovations could exacerbate existing inequalities, favoring large agro-industrial companies and marginalizing small-scale farmers and resource-constrained regions. Ensuring equitable access to CRISPR technology and its benefits is essential for promoting social justice and addressing global food security challenges (Levidow and Carr, 2020).
Furthermore, questions surrounding informed consent, transparency, and public engagement in decision-making processes related to CRISPR-modified crops are paramount. Stakeholder involvement, including farmers, consumers, policymakers, and civil society organizations, is crucial for fostering transparency, accountability, and democratic governance in agricultural biotechnology.
Conclusion
The advent of CRISPR-Cas technology heralds a new era in agriculture, offering unparalleled precision, efficiency, and adaptability in crop enhancement. With its transformative potential, CRISPR-Cas has emerged as a powerful tool for addressing the multifaceted challenges confronting contemporary agriculture. By leveraging the capabilities of CRISPR-Cas, researchers are poised to revolutionize crop breeding practices, enabling the development of resilient, high-yielding, and nutritionally enriched cultivars.
CRISPR-Cas-mediated genome editing facilitates the targeted modification of specific genes, thereby accelerating the breeding process and circumventing the limitations of traditional breeding methods (Zhang et al., 2019). This precision breeding approach holds tremendous promise for enhancing crop traits such as disease resistance, abiotic stress tolerance, and nutritional quality, thereby bolstering agricultural productivity and resilience in the face of climate change and environmental pressures (Wang et al., 2020). Furthermore, the versatility of CRISPR-Cas extends beyond genetic modification to encompass epigenome editing and gene regulation, offering novel avenues for crop improvement (Shimatani et al., 2017). By precisely modulating gene expression patterns and regulatory networks, CRISPR-Cas9 enables fine-tuning of agronomically important traits, such as flowering time, yield components, and nutrient utilization efficiency.
Moreover, CRISPR-Cas technology holds immense potential for promoting sustainable agriculture and addressing global food security challenges (Nalley et al., 2019). By enhancing crop productivity, reducing input requirements, and minimizing environmental impacts, CRISPR-edited crops offer a pathway towards achieving food security goals while mitigating the ecological footprint of agricultural production systems.
In conclusion, CRISPR-Cas technology represents a paradigm shift in agriculture, offering unprecedented opportunities for crop improvement and sustainable development. By harnessing the power of CRISPR-Cas, researchers can accelerate the pace of genetic improvement in crops, cultivate resilience to environmental stresses, and contribute to the realization of a food-secure future for generations to come.
References
Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan M Z, Ding S and Mahfouz M. 2018. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biology 19: 1. Doi: https://doi.org/10.1186/s13059-017-1381-1
Bao A, Chen H, Chen L, Chen S, Hao Q, Guo W, Qiu D, Shan Z, Yang Z, Yuan S, Zhang C, Zhang X, Liu B, Kong F, Li X, Zhou X, Tran L-S P and Cao D. 2019. CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biology 19: 131. Doi: https://doi.org/10.1186/s12870-019-1746-6
Bortesi L and Fischer R. 2015. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnology Advances 33(1): 41-52. Doi: https://doi.org/10.1016/j.biotechadv.2014.12.006
Čermák T, Baltes N J, Čegan R, Zhang Y and Voytas D F. 2015. High-frequency, precise modification of the tomato genome. Genome Biology 16(1): 1-14.
Choudhury P P, Singh R, Ghosh D and Sharma A R. 2016. Herbicide use in Indian agriculture (p. 110). ICAR - Directorate of Weed Research.
Chawla H S, Sodhi Y S and Arora S. 2017. CRISPR/Cas9 genome editing: are we there yet? Molecular Biotechnology 59(2-3): 241-255.
Chen K, Wang Y, Zhang R, Zhang H and Gao C. 2019. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annual Review of Plant Biology 70: 667–697. Doi: https://doi.org/10.1146/annurev-arplant-050718-100049
Chen Y, Wang Z, Ni H, Xu Y, Chen Q and Jiang L. 2017. CRISPR/Cas9-mediated base-editing system efficiently generates gain-offunction mutations in Arabidopsis. Science China Life Sciences 60: 520–523. Doi: https://doi.org/10.1007/s11427-017-9021-5
Christian M, Cermak T, Doyle E L, Schmidt C, Zhang F, Hummel A, Bogdanove A J and Voytas D F. 2010. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 186: 757–761. Doi: https://doi.org/10.1534/genetics.110.120717
Clasen B M, Stoddard T J, Luo S, Demorest Z L, Li J, Cedrone F, Tibebu R, Davison S, Ray E E, Daulhac A, Coffman A, Yabandith A, Retterath A, Haun W, Baltes N J, Mathis L, Voytas D F and Zhang F. 2016. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnology Journal 14(1): 169-176. Doi: https://doi.org/10.1111/pbi.12370
Doudna J A and Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213): 1258096. Doi: https://doi.org/10.1126/science.1258096
Eckerstorfer M F, Engelhard M, Heissenberger A, Simon S and Teichmann H. 2019. Plants developed by new genetic modification techniques - comparison of existing regulatory frameworks in the EU and non-EU countries. Frontiers in Bioengineering and Biotechnology 7: 26. Doi: 10.3389/fbioe.2019.00026
Feng j, Dai C, Luo H, Han Y, Liu Z, Kang C. 2019. Reporter gene expression reveals precise auxin synthesis sites during fruit and root development in wild strawberry. Journal of Experimental Botany 70(2): 563–574. Doi: https://doi.org/10.1093/jxb/ery384
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang D-L, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X and Zhu J-K. 2014. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/cas-induced gene modifications in arabidopsis. Procedings of National Academy of Science, USA. 111: 4632–4637. Doi: https://doi.org/10.1073/pnas.1400822111
Fister A S, Landherr L, Maximova S N, Guiltinan M J. 2018. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Frontiers in Plant Science 9: 268. Doi: https://doi.org/10.3389/fpls.2018.00268
Gasiunas G, Barrangou R, Horvath P and Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences 109(39): E2579-E2586.
Gumtow R, Wu D, Uchida J and Tian M A. 2018. Phytophthora palmivora extracellular cystatin-like protease inhibitor targets papain to contribute to virulence on papaya. Molecular Plant Microbe Interactions 31: 363–373 Doi: 10.1094/MPMI-06-17-0131-FI
Hsu P D, Lander E S and Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157(6): 1262-1278.
Hummel A W, Chauhan R D, Cermak T, Mutka A M, Vijayaraghavan A, Boyher A, Starker C G, Bart R, Voytas D F and Taylor N J. 2018. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnology Journal 16: 1275–1282.
Jia H and Wang N. 2014. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9(4): e93806.
Jia H, Orbović V and Wang N. 2017. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnology Journal 15(12): 1509-1519.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna J A and Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816-21. doi:10.1126/science.1225829
Kang B-C, Woo J W, Kim S-T, Bae S-J, Choi M, Kim J-S and Kim S-G. 2019. Guidelines for C to T base editing in plants: base-editing window, guide RNA length, and efficient promoter. Plant Biotechnology Reports 13: 533-541. Doi: https://doi.org/10.1007/s11816-019-00572-x
Kapusi E, Corcuera-Gómez M, Melnik S, Stoger E. 2017. Heritable Genomic fragment deletions and small indels in the putative ENGase gene induced by CRISPR/Cas9 in barley. Frontiers in Plant Science 8: 540. Doi: http://doi.org/10.3389/fpls.2017.00540
Kim H, Kim S-T, Ryu J, Kang B-C, Kim J-S, Kim S-G. 2017. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nature Communication 8: 14406. Doi: https://doi.org/10.1038/ncomms14406
Klap C, Yeshayahou E, Bolger A M, Arazi T, Gupta S K, Shabtai S, Usadel B, Salts Y and Barg R. 2017. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnology Journal 15: 634–647. Doi: https://doi.org/10.1111/pbi.12662
Lassoued, Macall D M, Smyth S J, Peter W B, Phillips, Hesseln H. 2020. How should we regulate products of new breeding techniques? Opinion of surveyed experts in plant biotechnology. Biotechnology Reports 26: e00640 Doi: https://doi.org/10.1016/j.btre.2020.e00460
Levidow L and Carr S. 2020. Governing agricultural sustainability in an era of biotechnologies and bioeconomies: Coordination, practices, and future challenges. Agriculture and Human Values 37(2): 363-375.
Li R, Zhang L, Wang L, Chen L, Zhao R, Sheng J and Shen L. 2018. Reduction of Tomato-Plant Chilling Tolerance by CRISPR–Cas9-Mediated SlCBF1 Mutagenesis. Journal of Agricultural and Food Chemistry 66(34): 9042-9051
Li T, Liu B, Spalding M H, Weeks D P and Yang B. 2012. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nature Biotechnology 30(5): 390-392.
Li Y, Zhu J, Wu H, Liu C, Huang C, Lan J, Zhao Y and Xie C. 2020. Precise base editing of non-allelic acetolactate synthase genes confers sulfonylurea herbicide resistance in maize. The Crop Journal 8(3): 449-456
Lu H-P, Luo T, Fu H-W, Wang L, Tan –Y, Huang J-Z, Wang Q, Ye G-Y, Gatehouse A M R, Lou Y-G and Shu Q-Y. 2018. Resistance of Rice to Insect Pests Mediated by Suppression of Serotonin Biosynthesis. Nature Plants 4: 338–344. Doi: https://doi.org/10.1038/s41477-018-0152-7
Ludwig Y, Dueñas C Jr, Arcillas E, Macalalad-Cabral R J, Kohli A, Reinke R, Slamet-Loedin I H. 2024. CRISPR-mediated promoter editing of a cis-regulatory element of OsNAS2 increases Zn uptake/translocation and plant yield in rice. Frontiers in Genome Editing 5:1308228. Doi: 10.3389/fgeed.2023.1308228
Jia H, Zhang Y, Orbovi´c V, Xu J, White F F, Jones J B and Wang N. 2017. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnology Journal 15: 817–823.
Ma J, Chen J, Wang M, Ren Y, Wang S, Lei C and Cheng Z. 2018. Sodmergen, null disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. Journal of Experimental Botany 69: 1051–1064. Doi: http://doi.org/10.1093/jxb/erx458
Malnoy M, Viola R, Jung M-H, Koo O-J, Kim S, Kim J-S, Velasco R, Kanchiswamy N C. 2016. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Frontiers in Plant Science 7: 1904.
Martín-Pizarro C, Triviño J C, Posé D. 2019. Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. Journal of Experimental Botany 70(3):885-895. doi: 10.1093/jxb/ery400
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H and Qu L-J. 2013. Targeted mutagenesis in rice using CRISPR-cas system. Cell Research 23: 1233–1236. Doi: https://doi.org/10.1038/cr.2013.123
Mishra R, Joshi R K and Zhao K. 2018. Genome editing in rice: Recent advances, challenges, and future implications. Front. Plant Science 9. Doi: http://doi.org/10.3389/fpls.2018.01361
Nalley L L, Tsiboe F and Durand-Morat A. 2019. The economic potential for CRISPR/Cas9 technologies in weed management. Crop Protection 119: 105-111.
Nekrasov V, Wang C, Win J, Lanz C, Weigel D and Kamoun S. 2017. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Scientific Reports 7: 482
Nonaka S, Arai C, Takayama M, Matsukura C and Ezura H. 2017. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Scientific Reports 7: 5539196 Doi: 10.1038/s41598-017-06400-y
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia J C, Perez-Quintero A, Li T, Joon-Seob E, Chenhao Li, Hanna N, Bo Liu, Florence A, Coline S, Van T L, Gerbert S D, Sébastien C, Sarah M S , Inez H Slamet-Loedin , Casiana V C, Boris S, Wolf B F, Frank F W and Yang B. 2019. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology 37(11): 1344-1350.
Puchta H and Fauser F. 2014. Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant Journal 78(5): 727-741.
Puchta H. 2016. Using CRISPR/cas in three dimensions: Towards synthetic plant genomes, transcriptomes and epigenomes. Plant Journal 87: 5–15. Doi: https://doi.org/10.1111/tpj.13100
Sauer N J, Narváez-Vásquez J, Mozoruk J, Miller R B, Warburg Z J, Woodward M J, Mihiret Y A, Lincoln T A, Segami R E, Sanders S L, Walker K A, Beetham P R, Schopke C R, Gocal G F W. 2016. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiology 170: 1917–1928. Doi: http://doi.org/10.1104/pp.15.01696
Schindele P, Wolter F and Puchta H. 2018. Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Letters 592: 1954–1967. Doi: https://doi.org/10.1002/1873-3468.13073
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi J J, Qiu J-L and Gao C. 2013. Targeted genome modification of crop plants using a CRISPR-cas system. Nature Biotechnology 31: 686–688. Doi: https://doi.org/10.1038/nbt.2650
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, Ezura H, Nishida K, Ariizumi T and Kondo A. 2017. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nature Biotechnology 35(5): 441-443. Doi: 10.1038/nbt.3833
Svitashev S, Schwartz C, Lenderts B, Young J K, Cigan M A. 2016. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nature Communication 7: 13274. Doi: https://doi.org/10.1038/ncomms13274
Thomazella D, Brail Q, Dahlbeck D and Staskawicz B. 2016. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv Doi: https://doi.org/10.1101/064824
Tian S, Jiang L, Cui X, Zhang J, Guo S, Li M, Zhang H, Ren Y, Gong G, Zong M, Liu F, Chen Q and Xu Y. 2018. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Reports 37: 1353–1356. Doi: https://doi.org/10.1007/s00299-018-2299-0
Usman B, Nawaz G, Zhao N, Liao S, Liu Y, Li R. 2020. Precise Editing of the OsPYL9 Gene by RNA-Guided Cas9 Nuclease Confers Enhanced Drought Tolerance and Grain Yield in Rice (Oryza sativa L.) by Regulating Circadian Rhythm and Abiotic Stress Responsive Proteins. International Journal of Molecular Sciences 21(21): 7854. Doi: 10.3390/ijms21217854
Varkonyi-Gasic, Erika, Wang, Tianchi, Voogd, Charlotte, Jeon, Subin, Drummond, Revel S M, Gleave, Andrew P, Allan and Andrew C. 2018. Mutagenesis of kiwifruit CENTRORADIALIS -like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnology Journal 17(5): 869–880.
Veillet F, Perrot L, Chauvin L, Kermarrec M-P, Guyon-Debast A, Chauvin J-E, Nogué F, Mazier M. 2019. Transgene-Free Genome Editing in Tomato and Potato Plants Using Agrobacterium-Mediated Delivery of a CRISPR/Cas9 Cytidine Base Editor. International Journal of Molecular Sciences 20(2): 402. Doi: https://doi.org/10.3390/ijms20020402
Waltz E. 2016. CRISPR-edited crops free to enter market, skip regulation. Nature Biotechnology 34: 582. Doi: https://doi.org/10.1038/nbt0616-582
Waltz E. 2016. Gene-edited CRISPR mushroom escapes US regulation. Nature 532(7599): 293. Doi: https://doi.org/10.1038/nature.2016.19754
Waltz E. 2018. With a free pass, CRISPR-edited plants reach market in record time. Nature Biotechnology 36: 6–7. Doi: https://doi.org/10.1038/nbt0118-6b
Wan D-Y, Ye Guo Y, Cheng Y, Hu Y, Xiao S, Wang Y, Wen Y-Q. 2020. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Horticulture Research 7: 116. Doi: https://doi.org/10.1038/s41438-020-0339-8
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C and Qiu J L. 2014. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nature Biotechnology 32: 947–951. https://doi.org/10.1038/nbt.2969
Wang D, Zhang F and Gao G. 2020. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell 181: 136–150.
Wu J, Chen C, Xian G, Liu D, Lin L, Yin S, Sun Q, Fang Y, Zhang H, Wang Y. 2020. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnology Journal 18: 1857-1859 Doi: https://doi.org/10.1111/pbi.13368
Wurtzel E T, Vickers C E, Hanson A D, Millar A H and Cooper M. 2019. Synthetic biology in plastids. Plant Physiology 179(3): 901-919.
Xu R, Yang Y, Qin R, Li H, Qiu C, Li L, Wei P and Yang J. 2016. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. Journal of Genetics and Genomics 43: 529–532. Doi: https://doi.org/10.1016/j.jgg.2016.07.003
Zaidi S S, Mukhtar M S and Mansoor S. 2018. Genome Editing: Targeting Susceptibility Genes for Plant Disease Resistance. Trends in Biotechnology 36(9): 898-906
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N and Zhu J K. 2019. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnology Journal 17(2): 225-238.
Zhang R, Chen S, Meng X, Chai Z, Wang D, Yuan Y, Chen K, Jiang L, Li J and Gao C. 2021. Generating broad-spectrum tolerance to ALS-inhibiting herbicides in rice by base editing. Science China Life Sciences 64: 1624–1633. https://doi.org/10.1007/s11427-020-1800-5
Zhang R, Liu J, Chai Z, Chen S, Bai Y, Zong Y, Chen K, Li J, Jiang L and Gao C. 2019. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nature Plants 5: 480-485
Zhang S, Wang L, Zhao R, Yu W, Li R, Li Y, Sheng J, and Shen L. 2018. Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants. Journal of Agricultural and Food Chemistry 66(34): 8949-8956. Doi: 10.1021/acs.jafc.8b02191
Zhang Y, Bai Y, Wu G, Zou S, Chen Y, Gao C and Tang D. 2017. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant Journal 91: 714–724. Doi: http://doi.org/10.1111/tpj.13599
Zhou H, He M, Li J, Chen L, Huang Z, Zheng S, Zhu L, Ni E, Jiang D, Zhao B and Zhuang C. 2016. Development of commercial thermo-sensitive genetic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-Mediated TMS5 editing system. Scientific Reports 6: 37395. Doi: https://doi.org/10.1038/srep37395
Copyright
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.

